Creating GO Annotations In Noctua
Creating a basic Activity Unit (MF, part of BP, occurs in CC)
In this hands-on tutorial, you will learn how to create a basic activity using C. elegans as an example:
1. Open the Activity Unit Form
- Click on ‘Create Activity’ to select an Activity Unit Form
- A new empty form will slide in from left side
2. Add a Gene Product
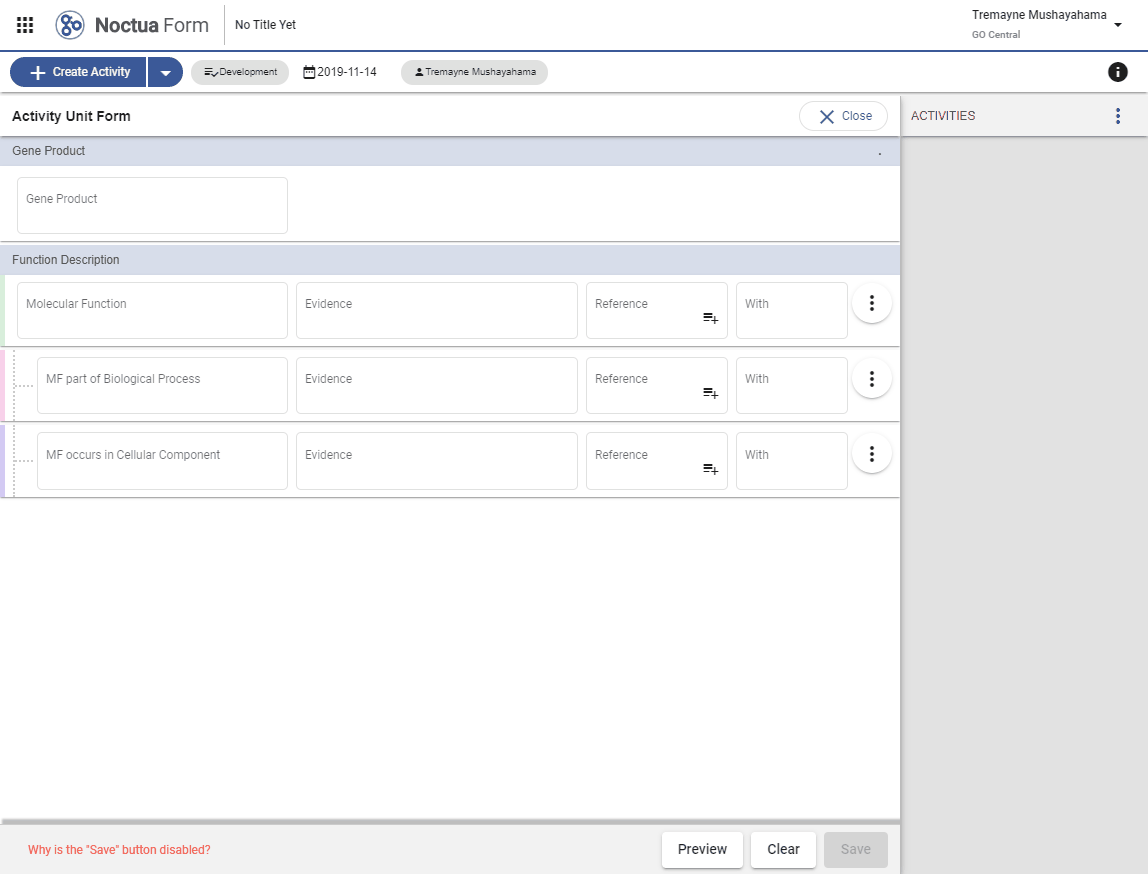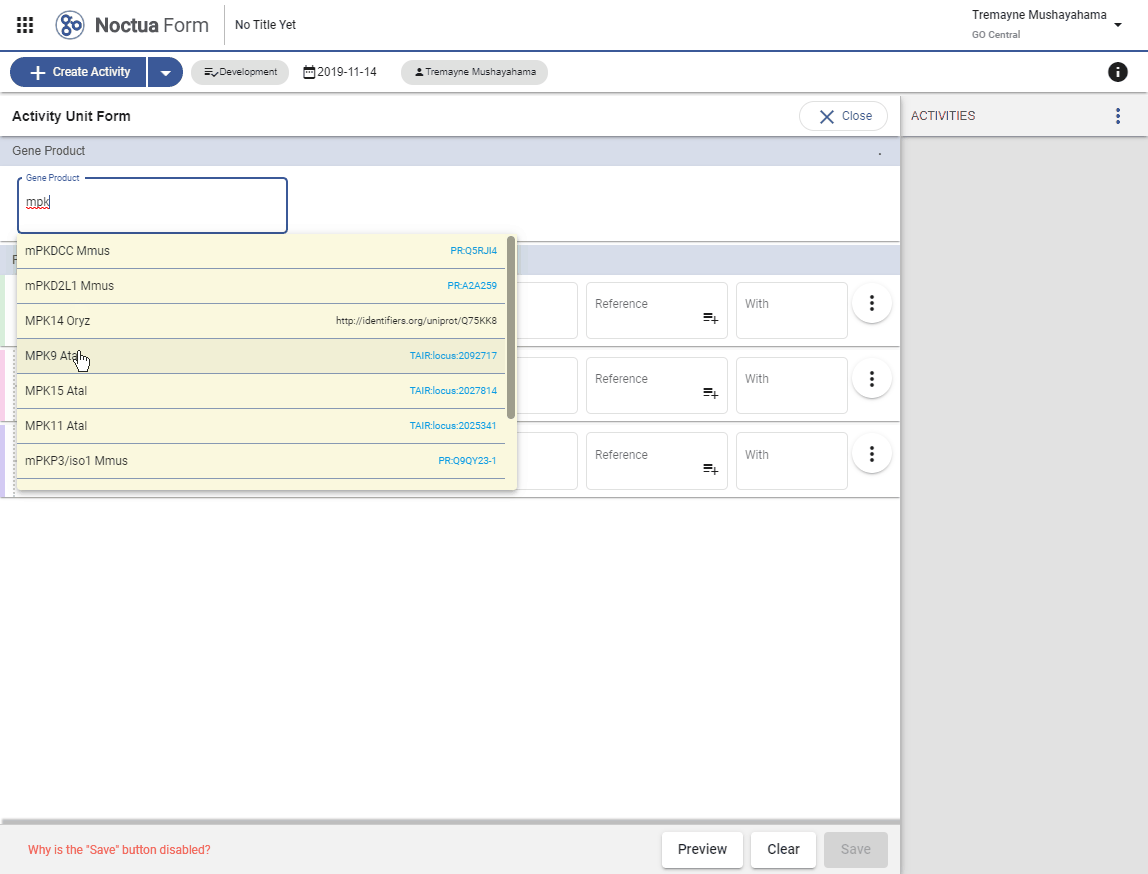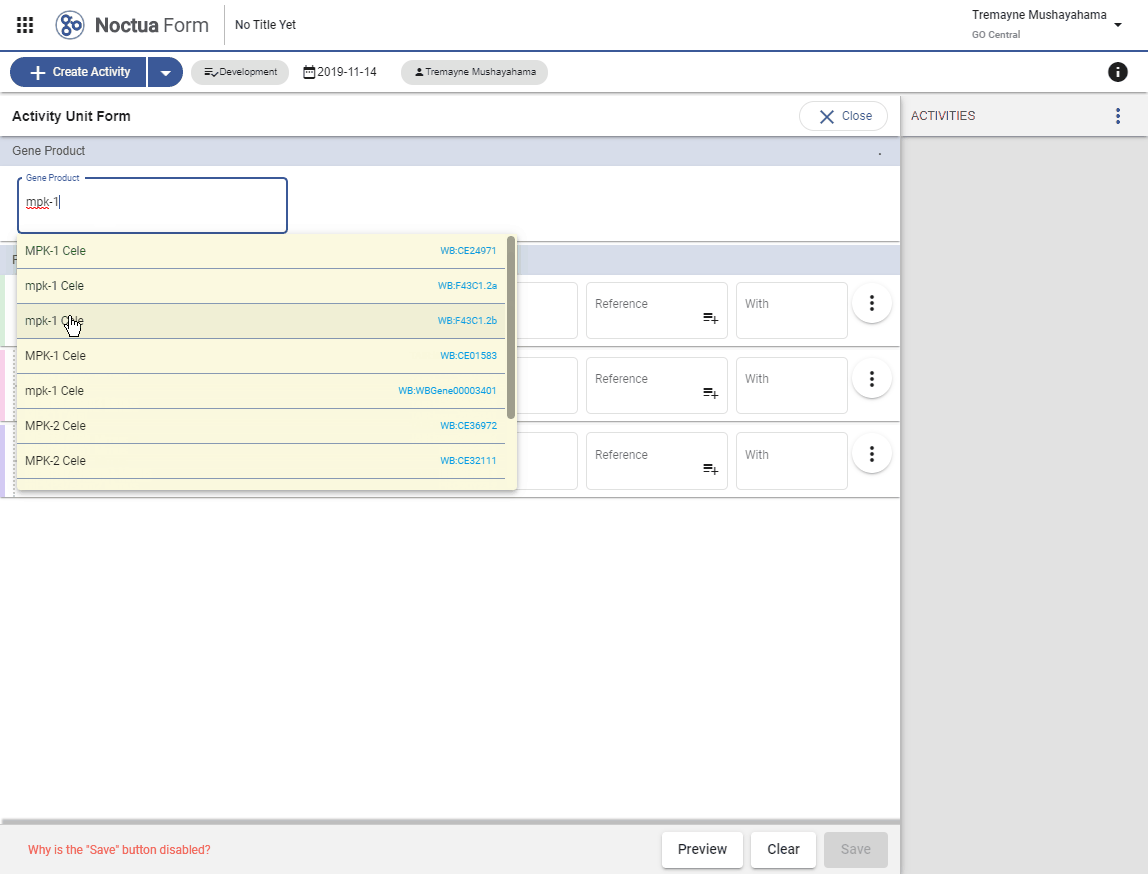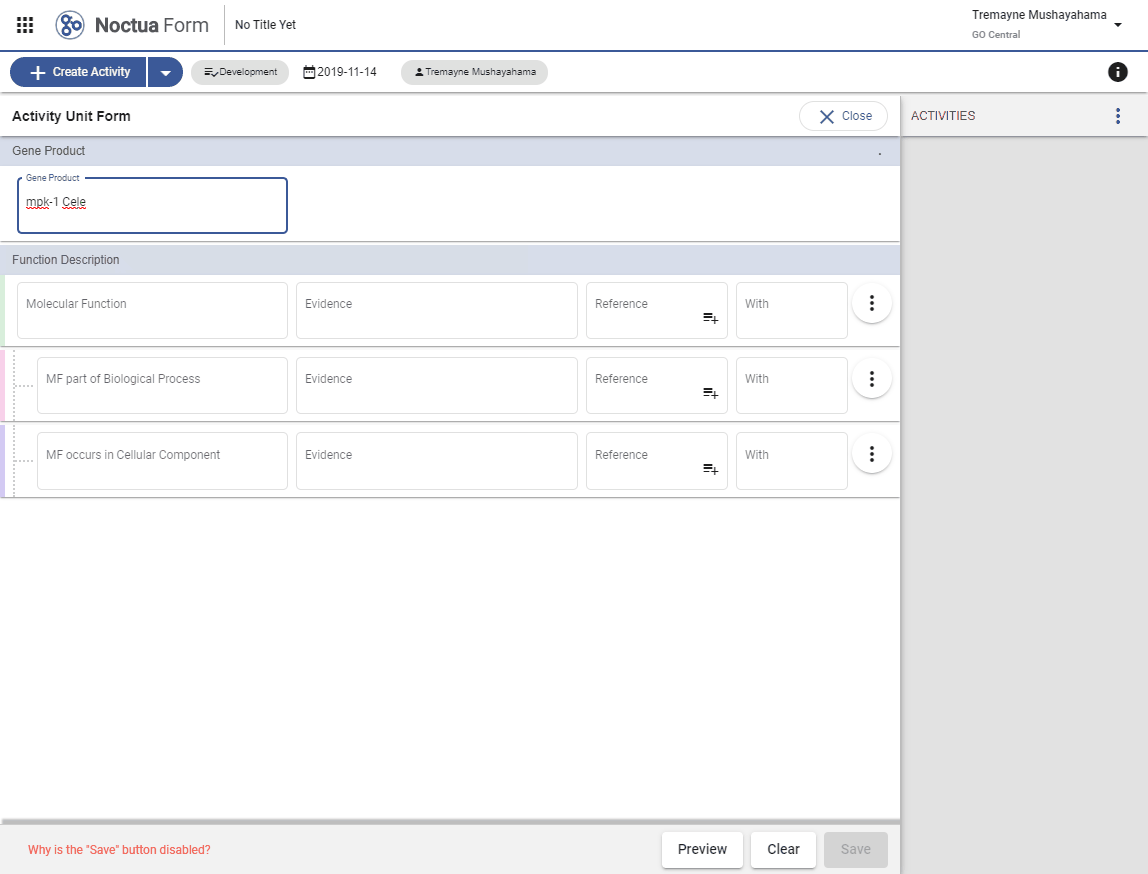
- Type in gene name: mpk-1 (explain that returned entities are what is in gpi file and therefore in NEO)
- Autocomplete options are presented in a pop-up window
- Gene products link out to respective gene product pages via dbxrefs.yaml
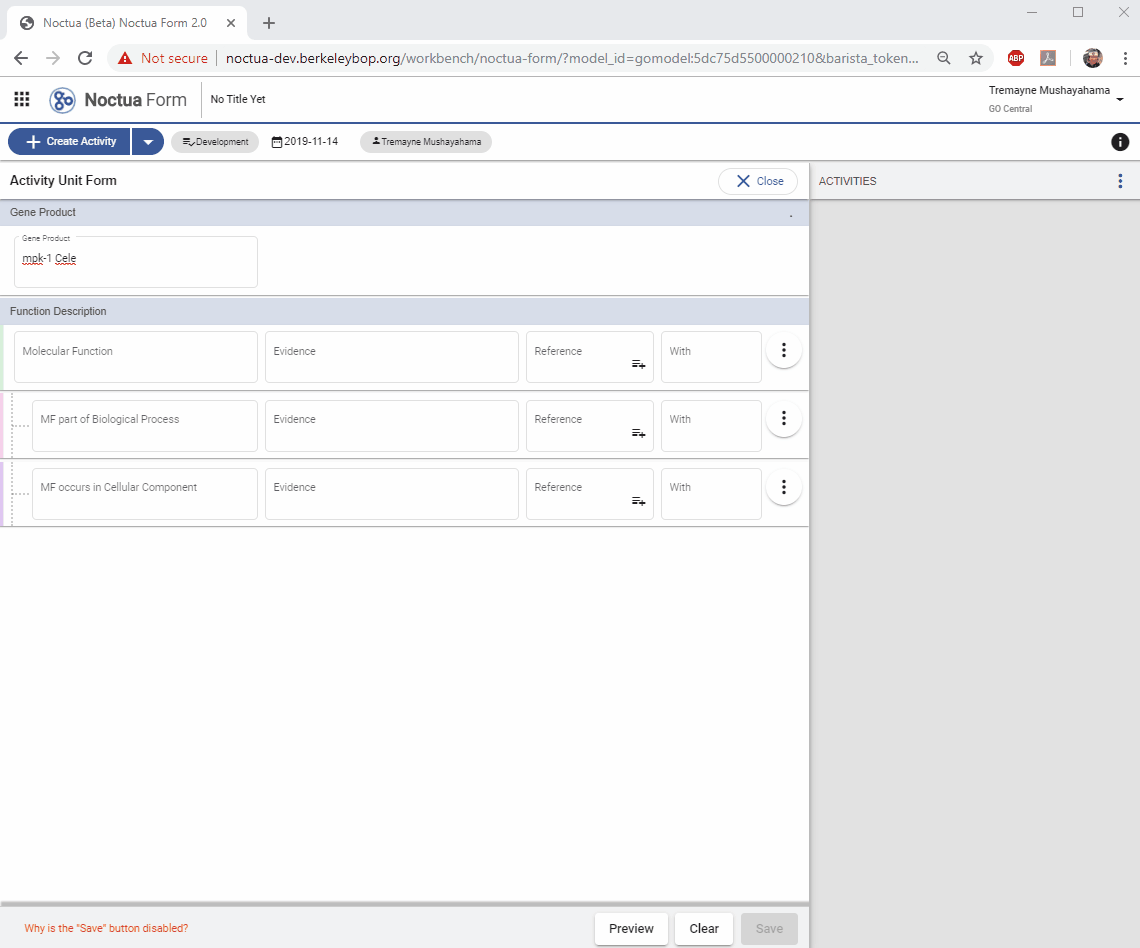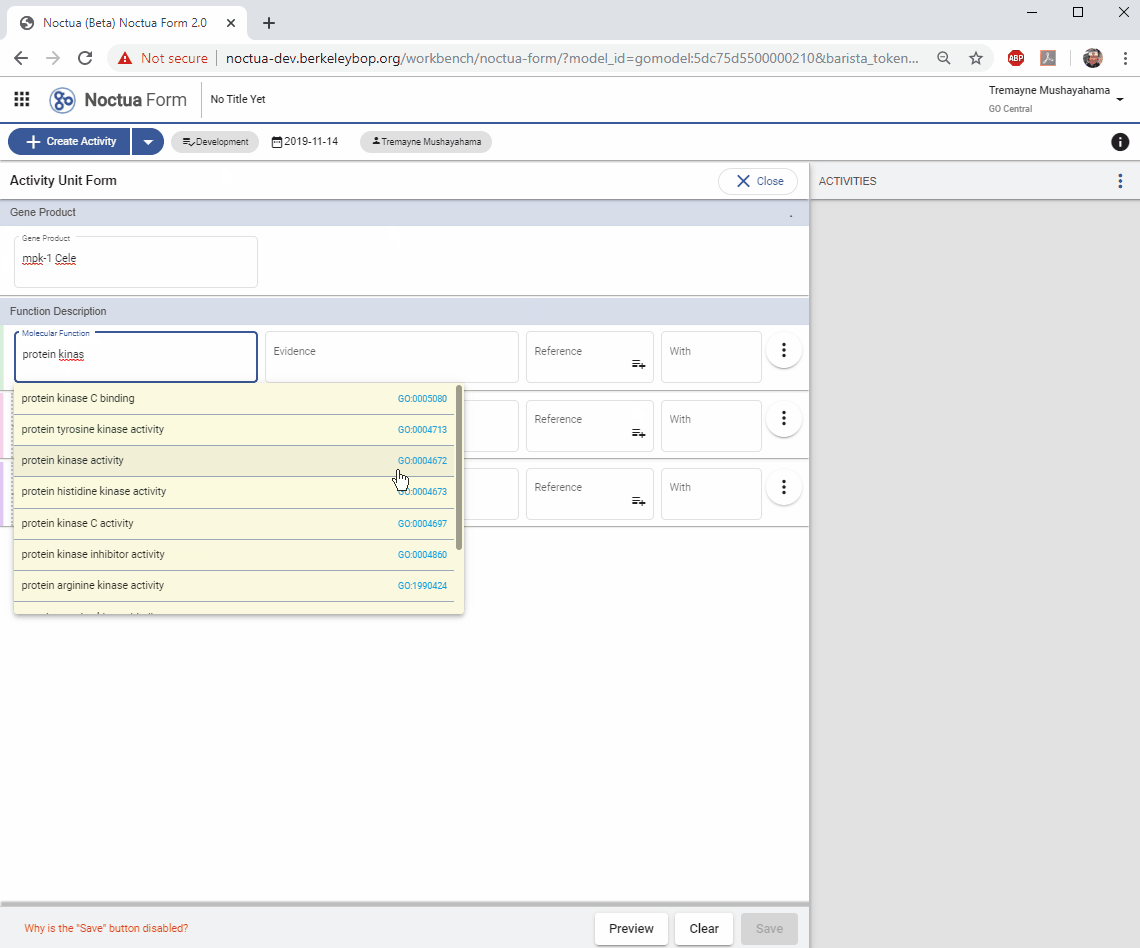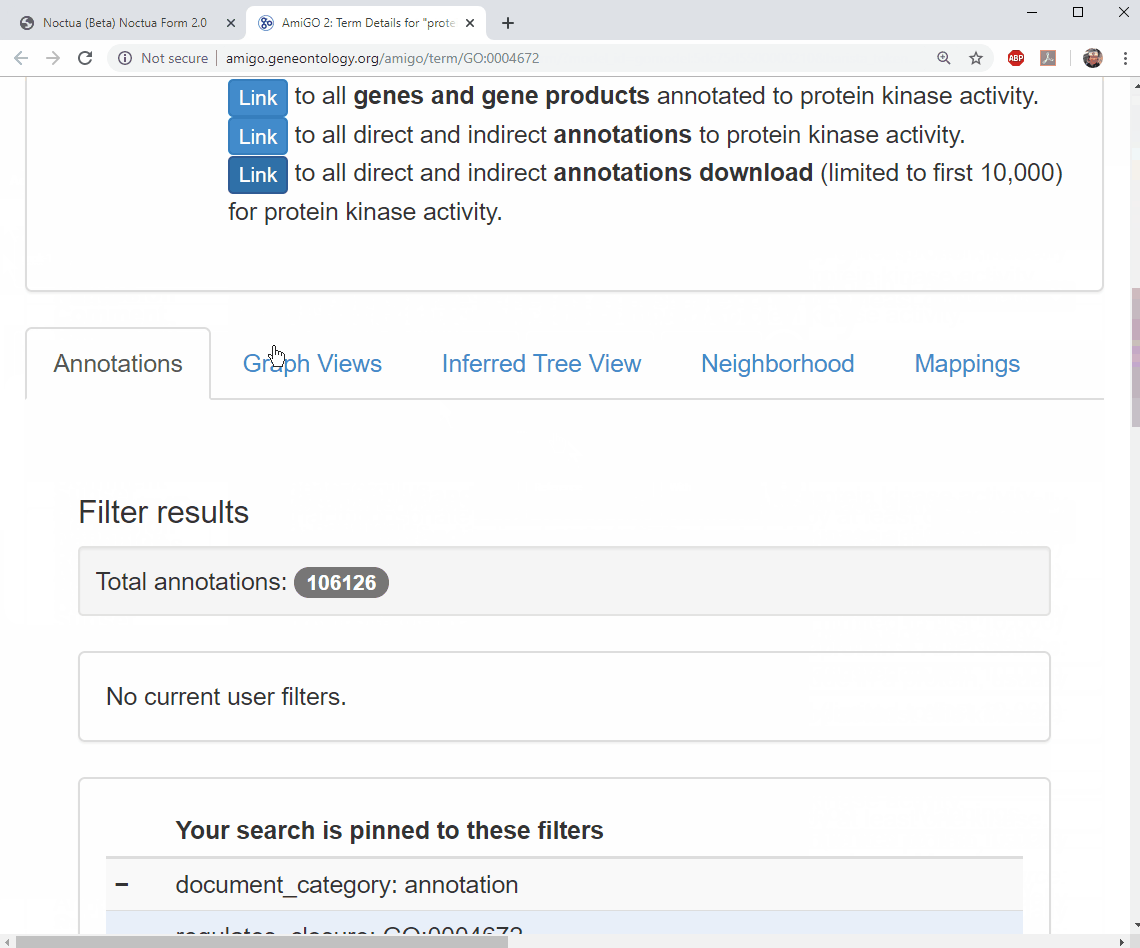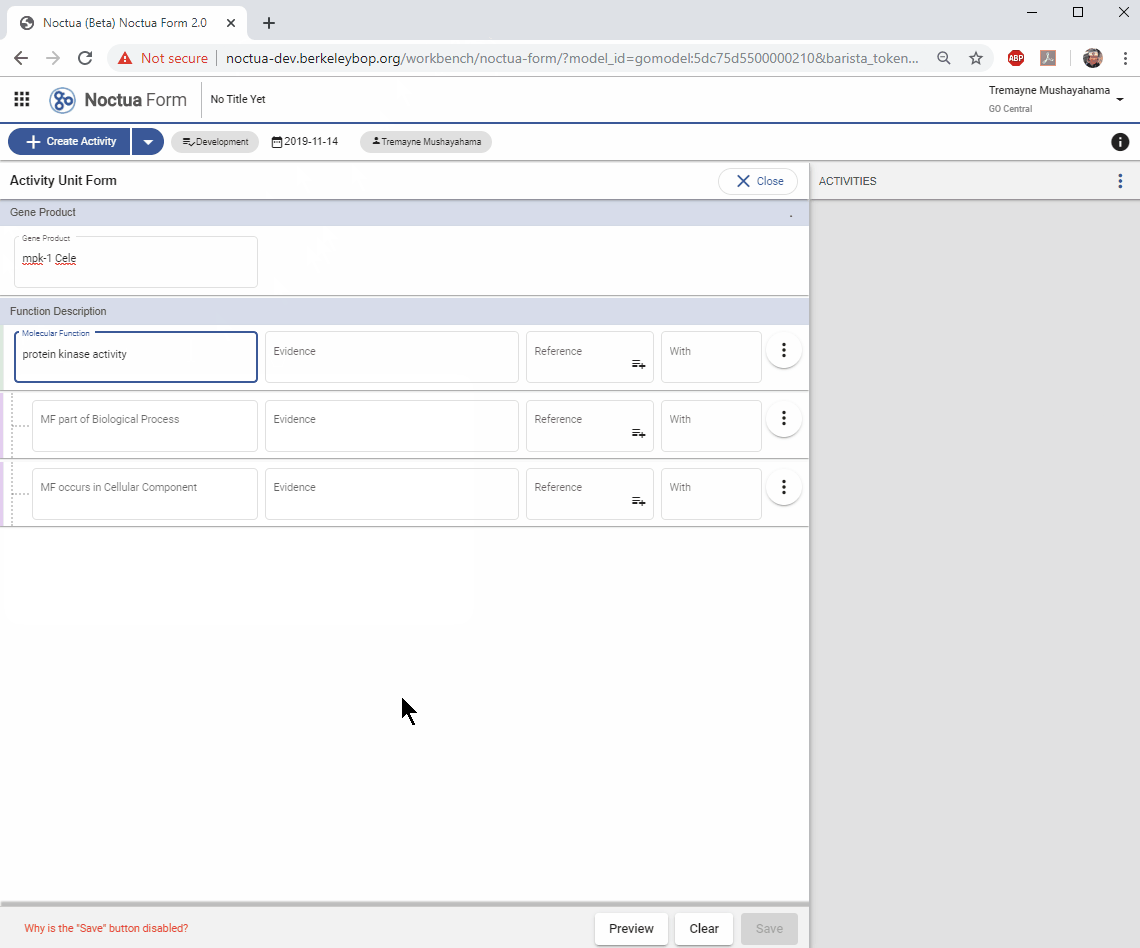
3. Add a Molecular Function
- Search for ‘protein kinase activity’
- Autocomplete options are presented in a pop-up window
- Terms link out to AmiGO
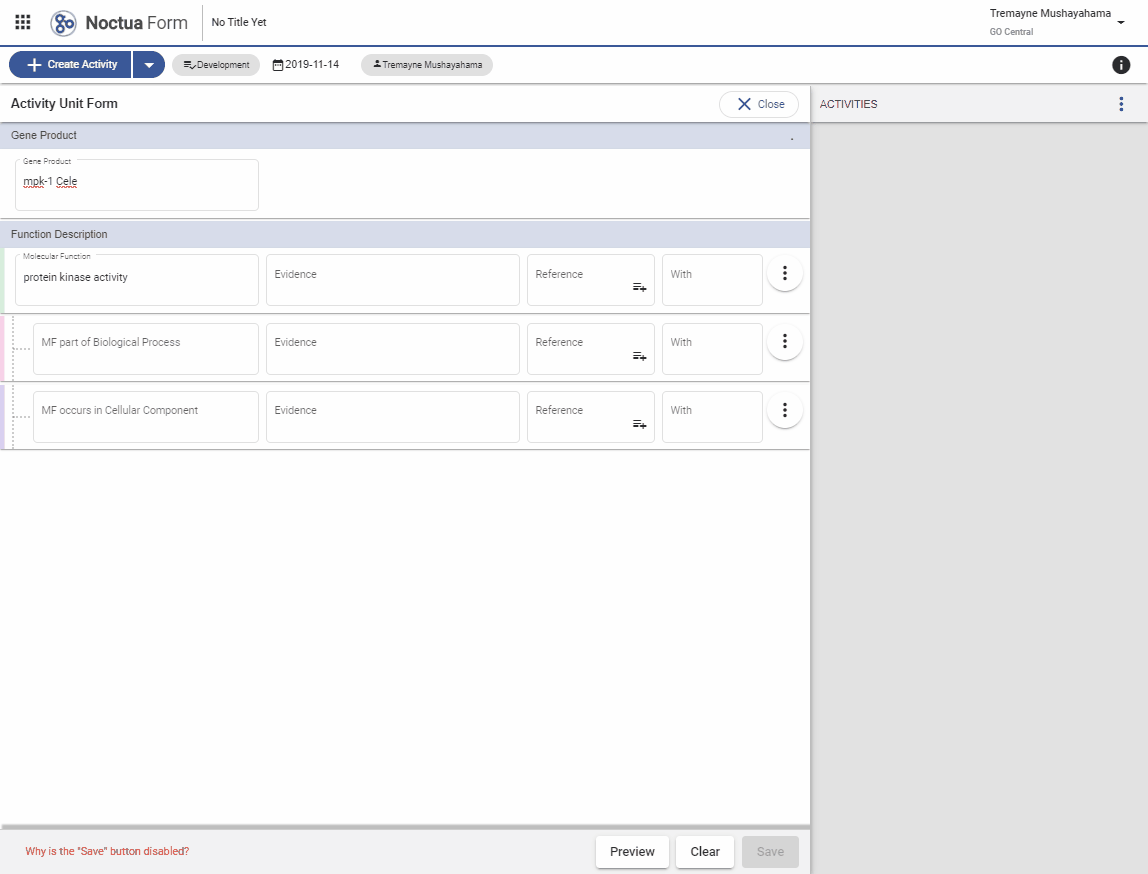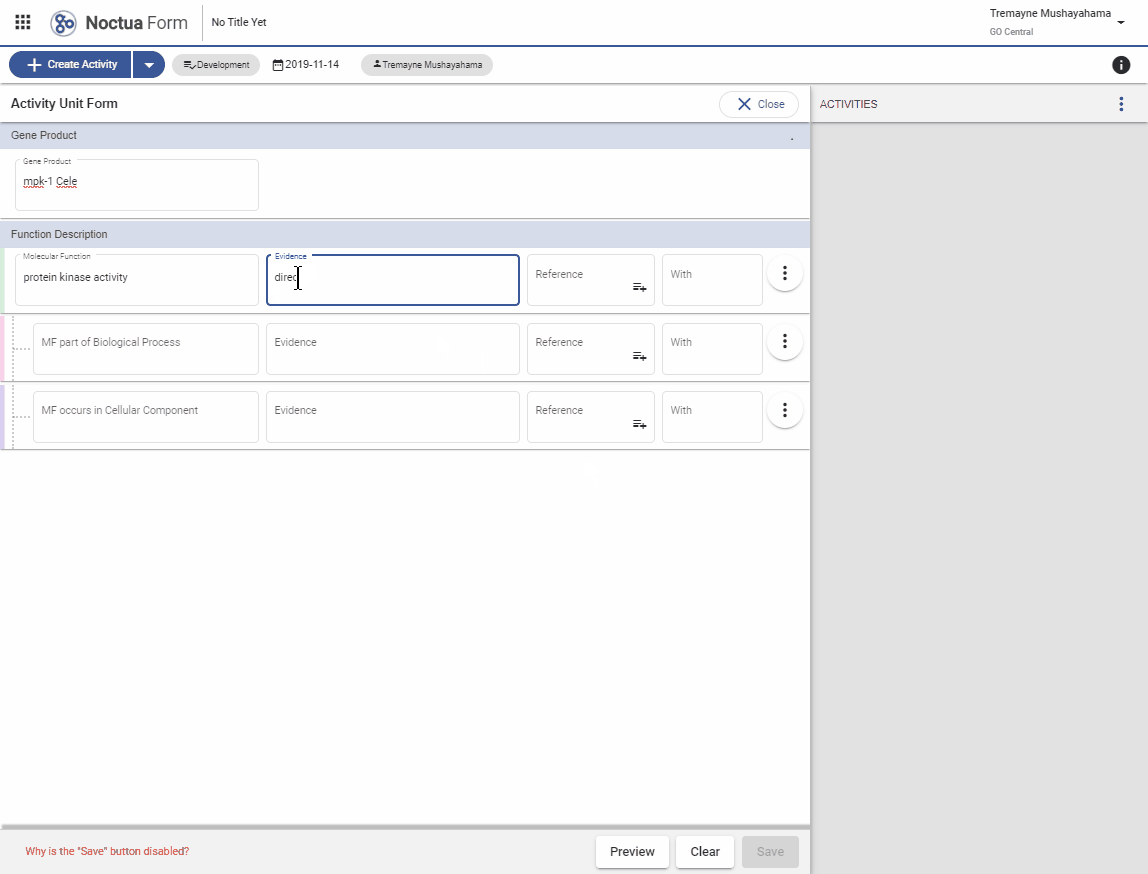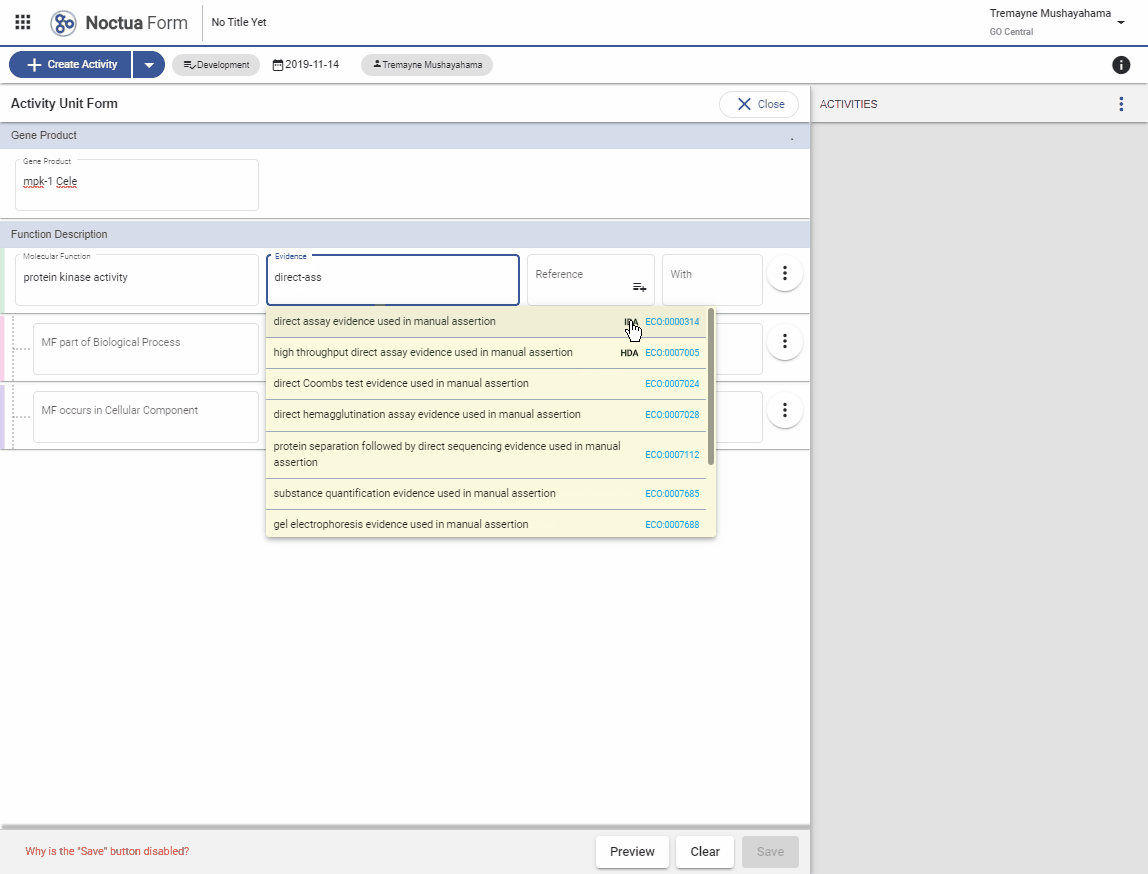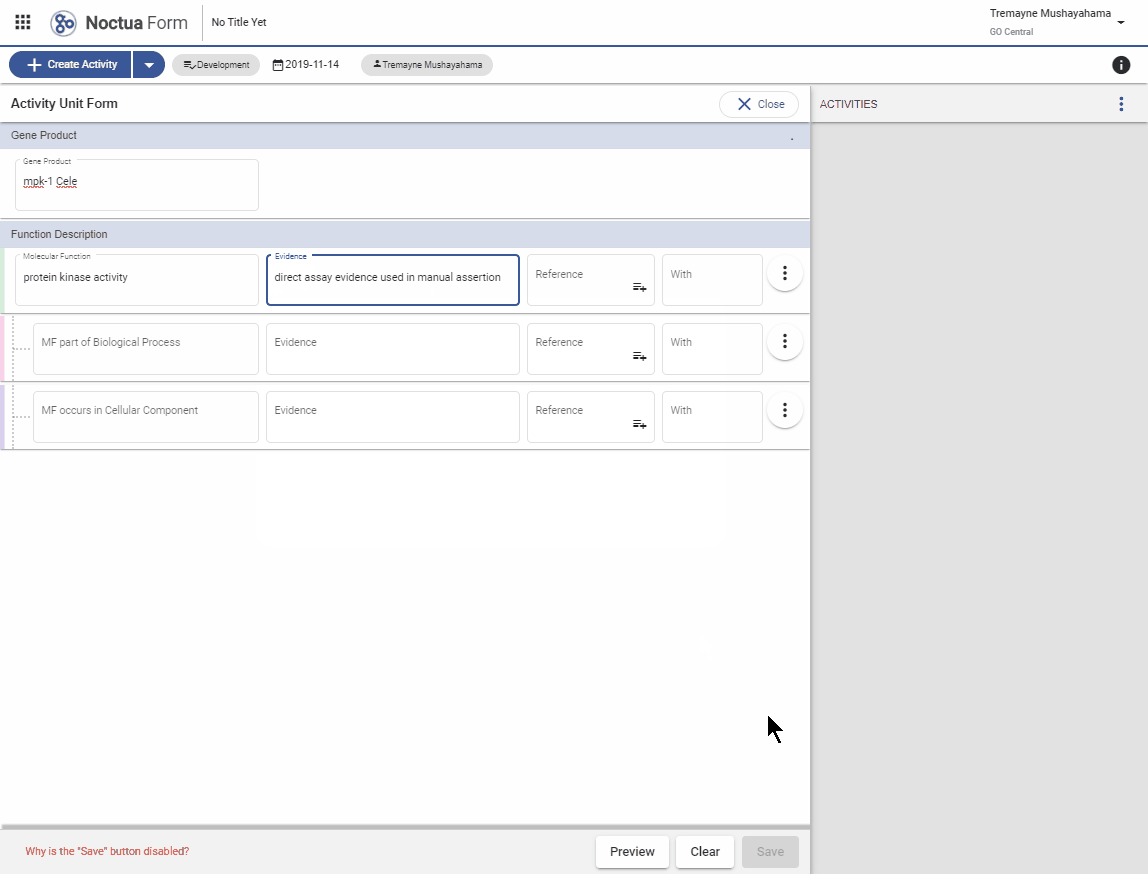
3.1. Evidence code autocomplete using ECO term names
- Search for ‘direct assay’
- GO codes are indicated with bold three-letter GO code
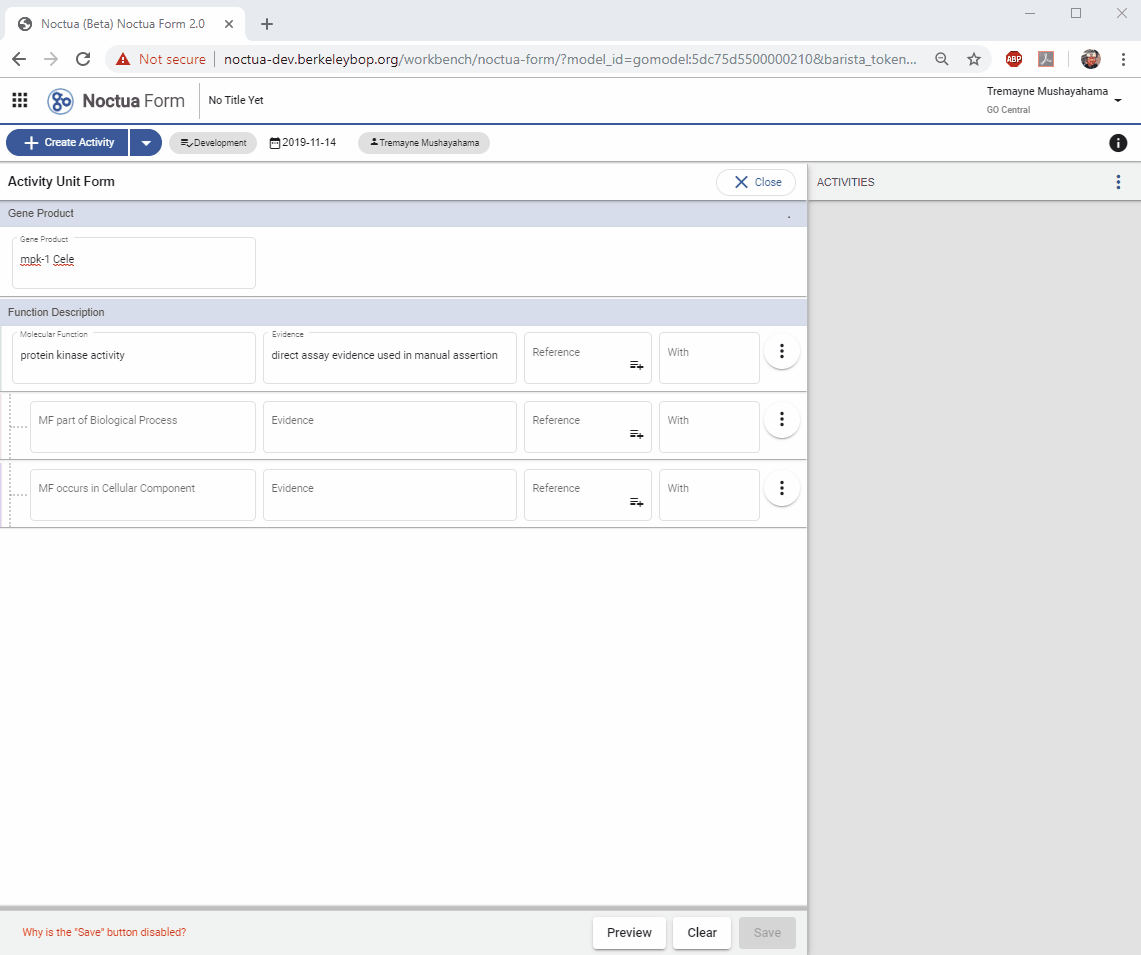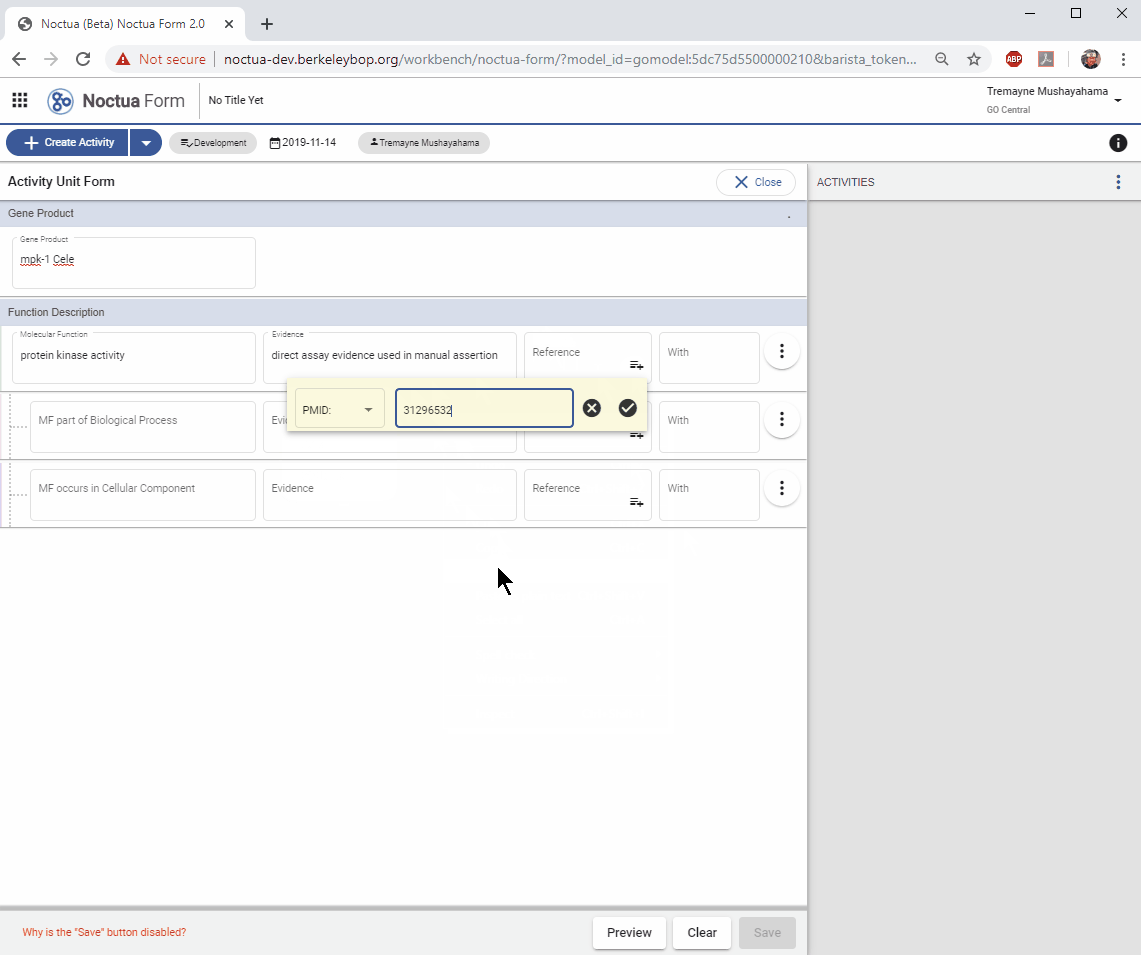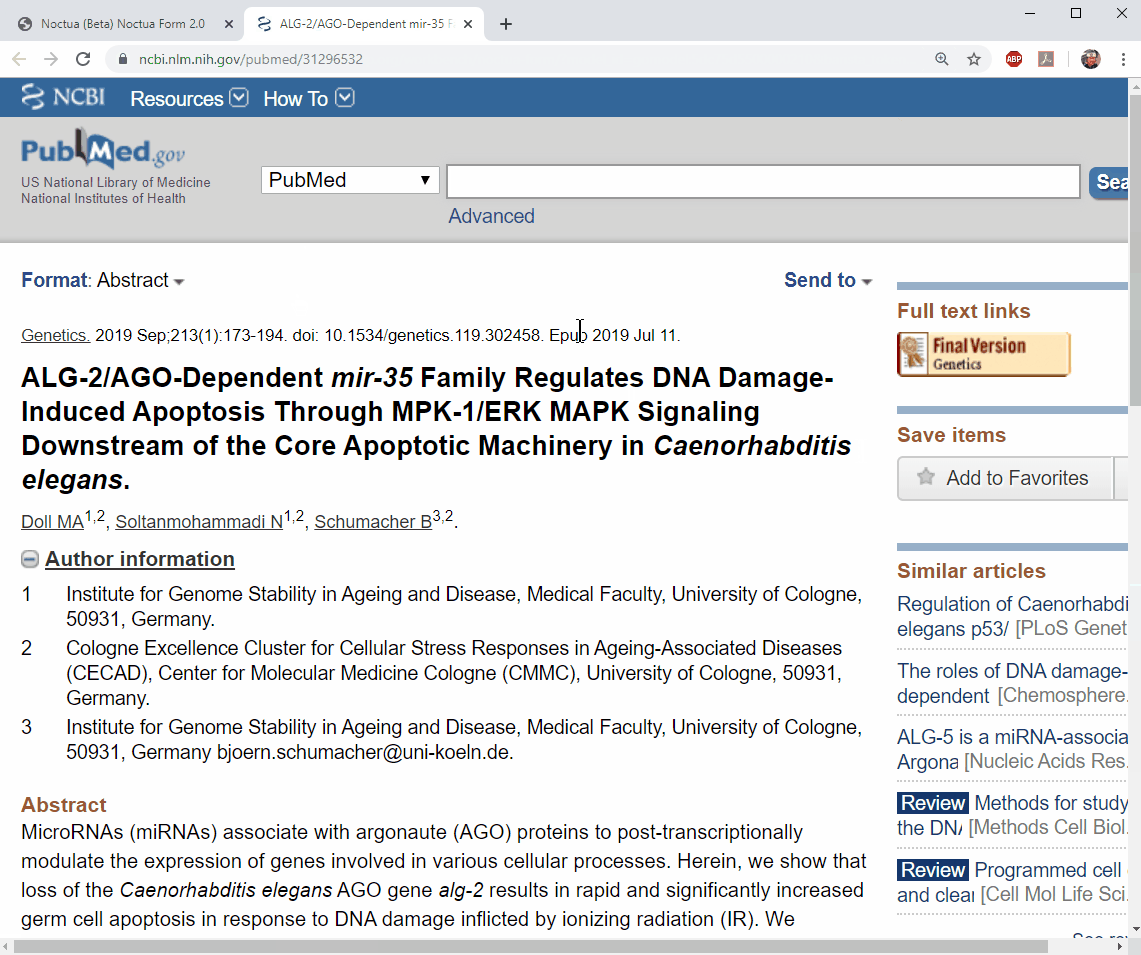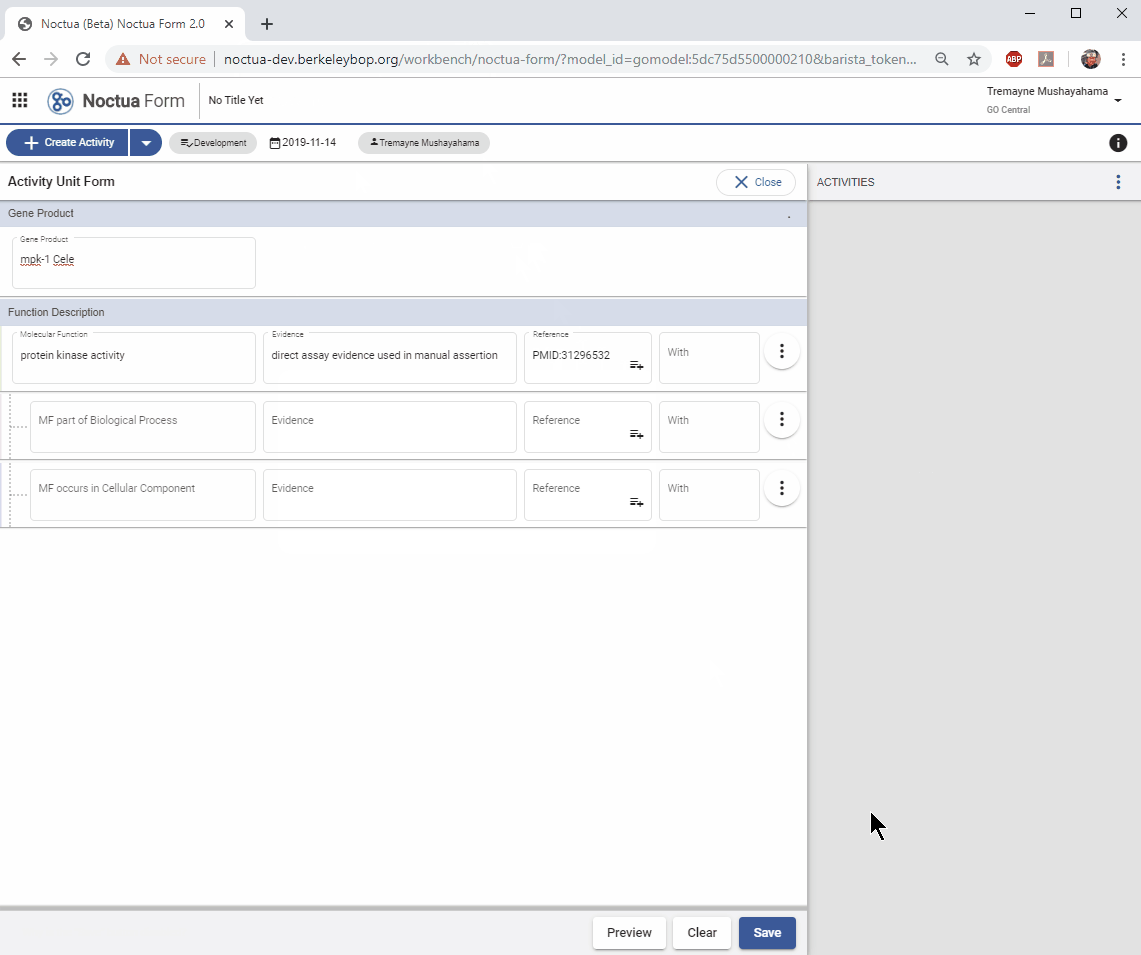
3.2. Add a reference. Two options:
- Cut and paste a DB and id, e.g. PMID:31296532 OR
- Click on the +/lines symbol
- Pop-up appears with a select drop-down of dbs for references: PMID,
- DOI, GO_REF
- Select PMID
- Add identifier: 31296532
Note that at this point you can save just your MF annotation by clicking on the SAVE button. This will save the MF annotation and automatically save the resulting model. However, if you want to add contextual information, such as MF inputs, or a Biological Process or Cellular Component annotation, you should wait until you’ve added all information before saving.
4. Add that MF is ‘part of’ a BP
- Search for ‘DNA damage response, detection of DNA damage’
- Autocomplete options are presented in a pop-up window
- Terms link out to AmiGO
- Evidence code autocomplete using ECO term names
- Search for ‘mutant phenotype’
- GO codes are indicated with bold three-letter GO code
- Add a reference. Two options:
- Cut and paste a DB and id, e.g. PMID:31296532 OR
- Click on the +/lines symbol
- Pop-up appears with a select drop-down of dbs for references: PMID, DOI, GO_REF
- Select PMID
- Add identifier: 31296532
5. Add that MF ‘occurs in’ a CC
- Search for ‘nucleus’
- Autocomplete options are presented in a pop-up window
- Terms link out to AmiGO
- Evidence code autocomplete using ECO term names
- Search for ‘direct assay’
- GO codes are indicated with bold three-letter GO code
- Add a reference. Two options:
- Cut and paste a DB and id, e.g. PMID:31296532 OR
- Click on the +/lines symbol
- Pop-up appears with a select drop-down of dbs for references: PMID, DOI, GO_REF
- Select PMID
- Add identifier: 31296532