Tools to browse, search, visualize and curate GO
The Gene Ontology provides a variety of tools to help users browse, search, visualize, download both the GO ontology and GO annotations.
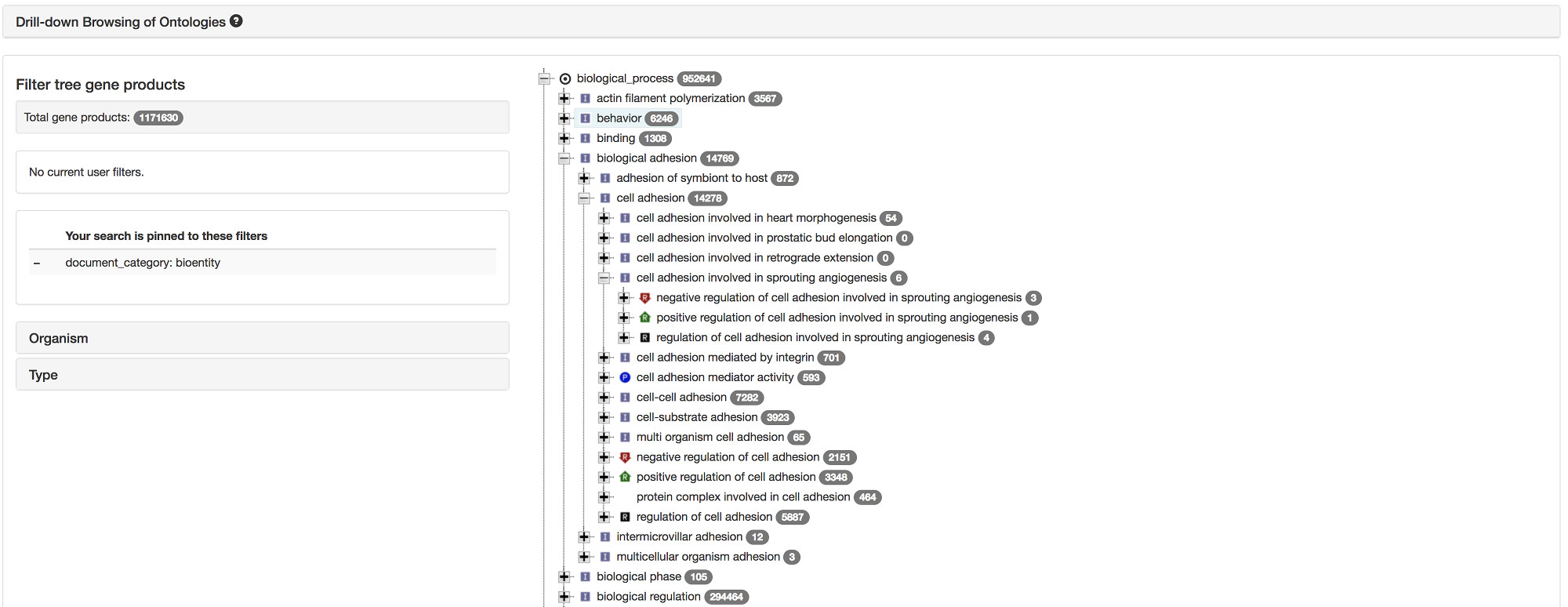
Browse the ontology
Drill-down browsing of the GO ontology is possible via an interactive tree.
To browse curated and inferred GO annotations, visit AmiGO.
Search the ontology and GO annotations
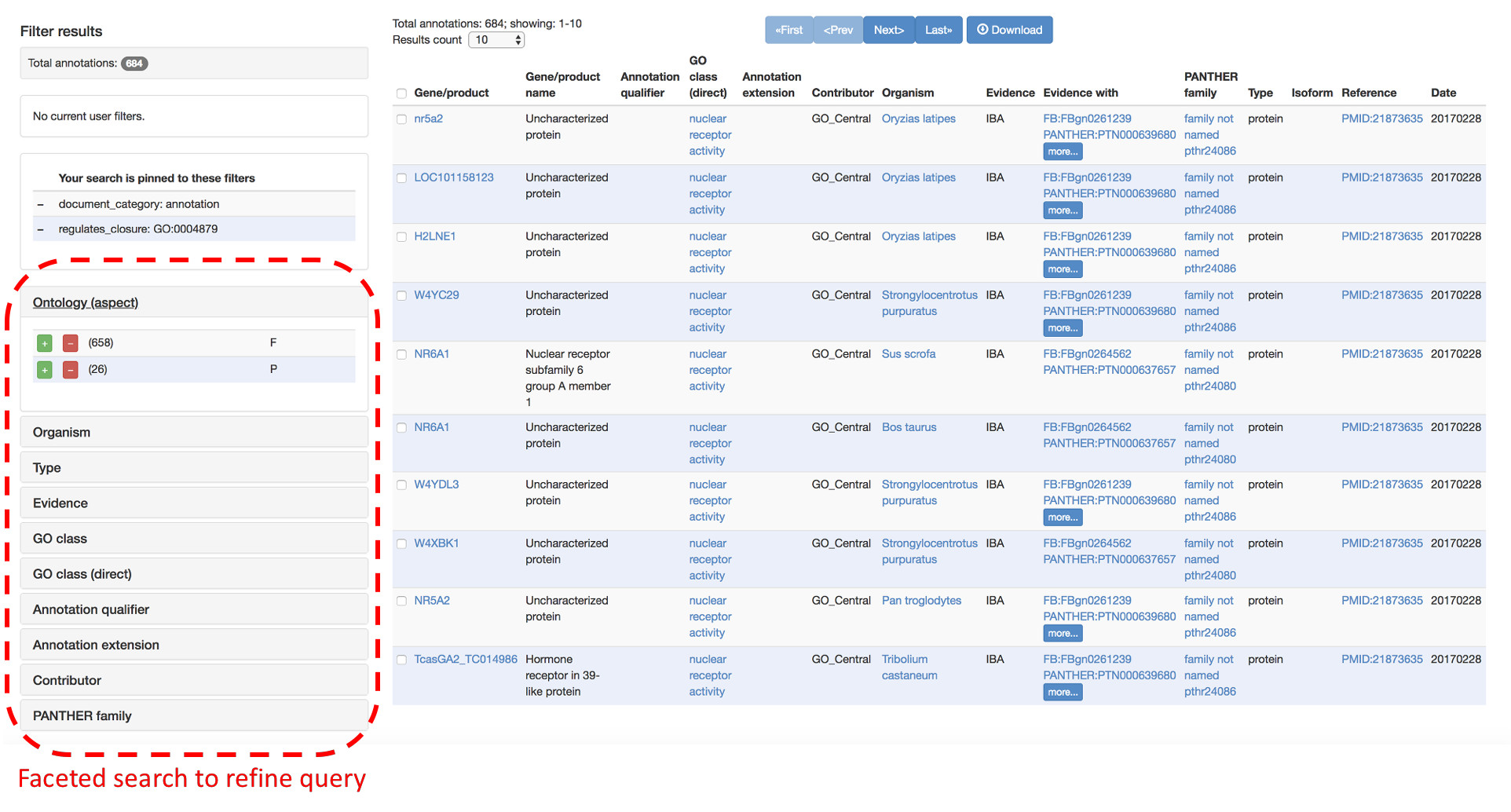
AmiGO can be used to search both the GO ontology, the GO annotations and details about gene products described in the GO knowledgebase.
AmiGO supports faceted search to refine queries by restricting specific parameters, such as a species, an ontology aspect (Biological Process, Molecular Function or Cellular Component), an evidence (e.g. only curated annotations), etc.
Visualize annotations with the GO Ribbon
Many genes have large numbers of annotations, making it difficult to get a quick overview of a gene function, or the functions of gene sets. GO ribbons are specifically designed to help users visualize and explore the functions of a gene. The ribbon is a lightweight, reusable widget that can be embedded in any website, and retrieves data directly from the GO knowledgebase via the GO API.
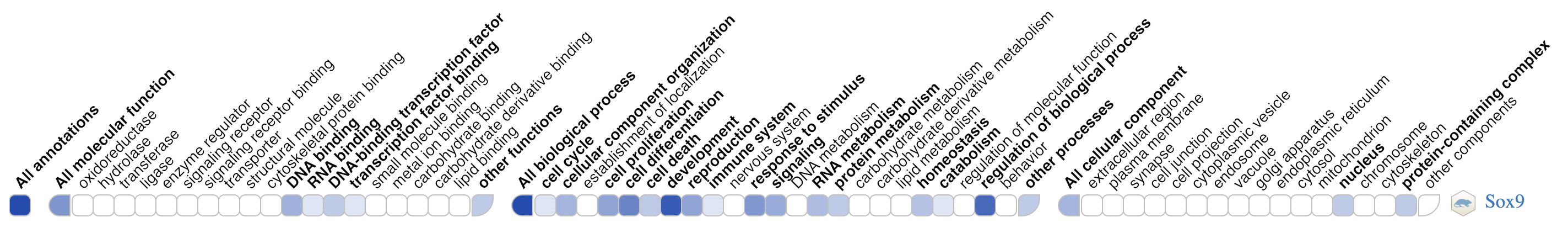
Interested to learn more about the GO Ribbon ? Check the GO Ribbon documentation
Want to try it now with your genes ? [ Access the GO Ribbon sandbox ]
The GO Ribbon is also integrated in the Alliance of Genome Resources and work is underway for further integration in several projects.
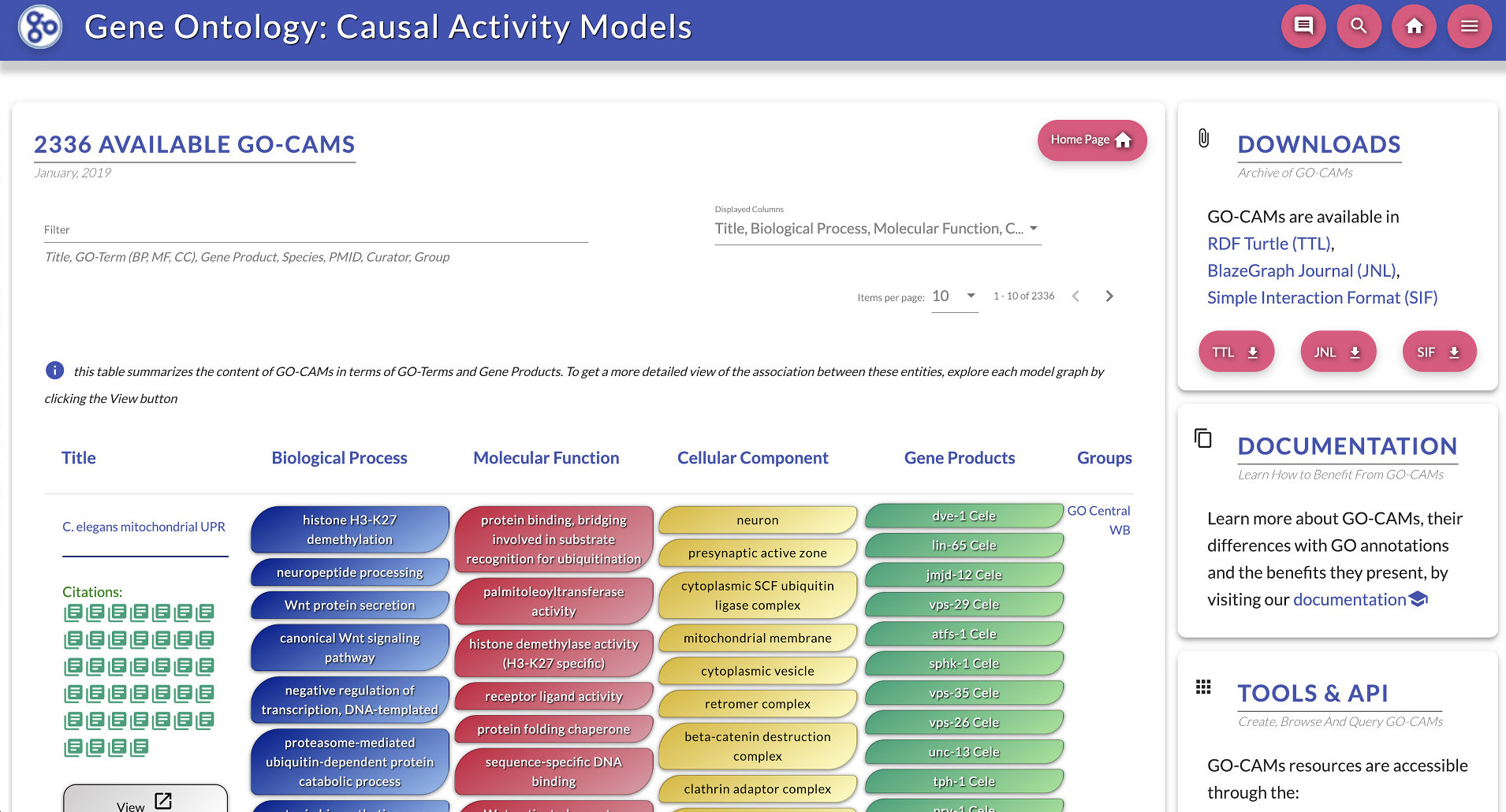
Search GO Causal Activity Models (GO-CAMs)
GO-Causal Activity Models (GO-CAMs) use a defined “grammar” for linking multiple standard GO annotations into larger models of biological function (such as “pathways”) in a semantically structured manner. GO-CAMs are created by expert biocurators from the GO Consortium, using the Noctua Curation Platform.
GO-CAMs can be browsed and searched in the http://geneontology.org/go-cam section of this site. For example, you can limit your search results to GO-CAMs having a certain GO term, a certain gene, or created by a specific curation group or curator.
Visualize GO-CAM Models with the Alliance of Genome Resources Pathway Viewer
Summaries of GO-CAM Models are viewable on the Alliance of Genome Resources webpages for relevant genes. The Alliance of Genome Resources Pathway Viewer is designed to easily visualize the pathways in which a gene product play a role, and to give easy access to the functions of the other genes in the same pathway, listed in a menu on the right (see example below).
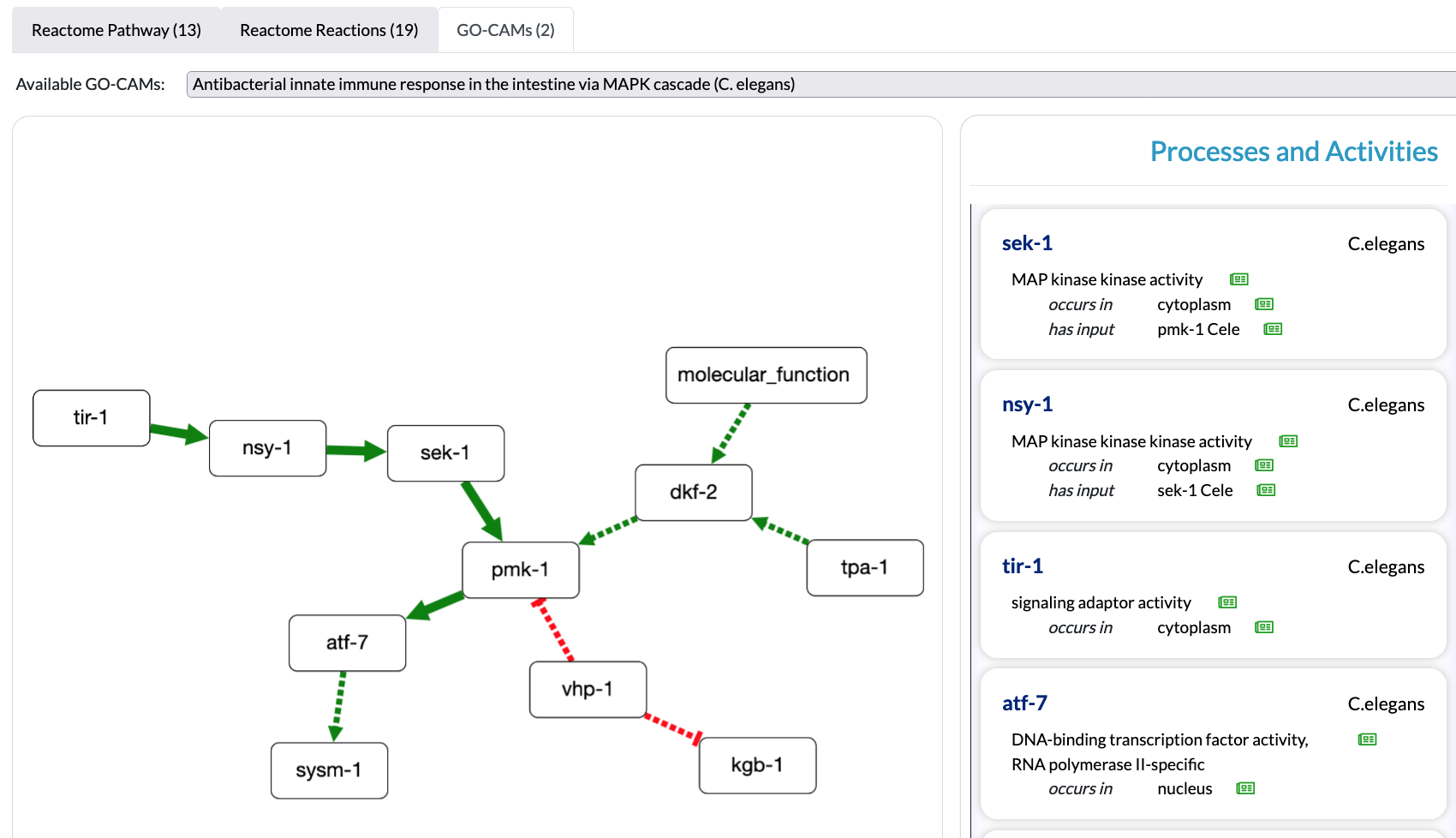
The Alliance of Genome Resources Pathway Viewer is integrated in the Alliance of Genome Resources and work is underway for further integration in several projects. Alliance of Genome Resources pages display models containing a minimum of 3 connected molecular functions (a.k.a. Activity Units).
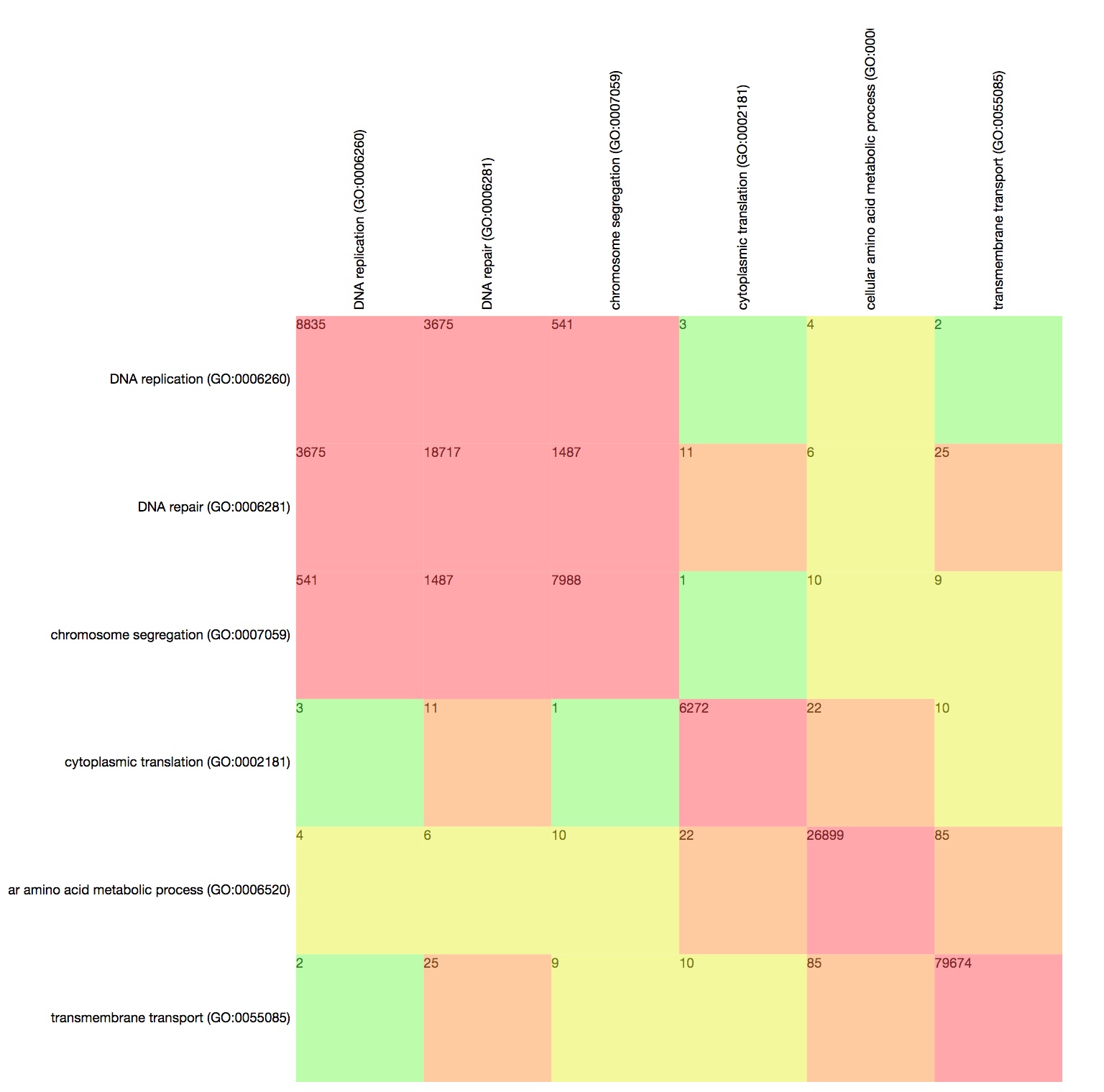
Explore co-annotations with the GO Term Matrix
The GO Term Matrix represents co-annotations of pairs of GO terms. It allows to compare multiple pairs of terms simultaneously.