Download annotations
Getting annotations for a selected organism
This page has instructions for getting GO annotations for almost any organism. If your organism is not available in the official GO products, UniProt GAFs by proteome, or NCBI RefSeq, we recommend using the latest version of InterProScan for unannotated organisms.
Jump to a section:
Required Files
Most tools that use GO annotations take two input files:
- a file with the annotations (in Gene Annotation Format, or GAF)
- a file with the GO ontology structure (in Open Biomedical Ontology Format, or OBO)
Because the ontology and annotations are constantly being improved over time, we recommend downloading the latest version of the annotations for your organism and the corresponding ontology file for that GO version. The version should be specified in the header of the annotation file.
Citing GO
To ensure reproducibility for any publication where GO was used at any point in the research, please include:
- appropriate GO publication(s)- refer to the full GO citation policy
- the URL where the files were obtained
- the date on the header of the GAF file
- the ontology version number
1. Commonly studied organisms
This GAF download page has annotations for selected commonly-studied species.
For organisms with many expert-curated GO annotations (those with MODs, dedicated databases, etc.), we recommend downloading annotations from the links in the above-linked table. These organisms often have a large number of manual annotations supported by direct experimental evidence as well as annotations based on other evidence types.
- These annotations should be used with the latest version of the GO ontology.
- Annotations for these organisms are also available as GPAD/GPI companion files; see the /annotations/ directory of the current release http://current.geneontology.org. For more information on these infrequently used filetypes see the format pages for GPAD+GPI.
2. All other organisms
For all other organisms we recommend downloading annotations from one of the following sources: UniProt or NCBI RefSeq. Both of these provide highly accurate computational methods. The header of the annotation file specifies the version of the ontology you should use to accompany the annotation file. Older versions of the GO ontology can be downloaded from the GO download archives.
- UniProt GAFs by proteome: Annotation files are available for about 20,000 complete proteomes (one protein sequence per protein-coding gene). Use these files if you want to use UniProtKB identifiers.
- Go to https://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/
- Navigate to your organism & download the
.goafile, e.g.22426.A_gambiae.goa
Tip: use your browser’s in-page search to find the species name.
- NCBI RefSeq: If your organism has a reference genome assembly in NCBI in the RefSeq collection (RefSeqs have assembly accessions starting with
GCF_), GO annotations are available in GAF format through NCBI Gene identifiers. Annotation files are available for all eukaryotic genomes available at NCBI RefSeq.Note: GO annotations are not currently available at NCBI for archaea, bacteria or viruses. GO annotations are not currently available at NCBI for eukaryotic genomes only in GenBank (only accession starts with
GCA_).- Start at the NCBI homepage
- Enter your organism in the search box near the top of the page and click Search, e.g. Anopheles gambiae
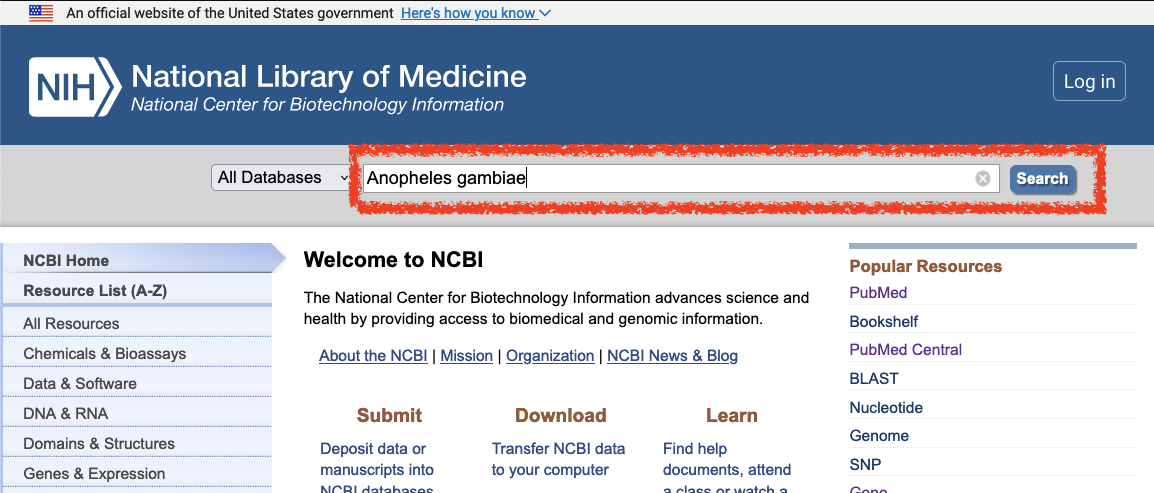
- Follow the “Genomes” link
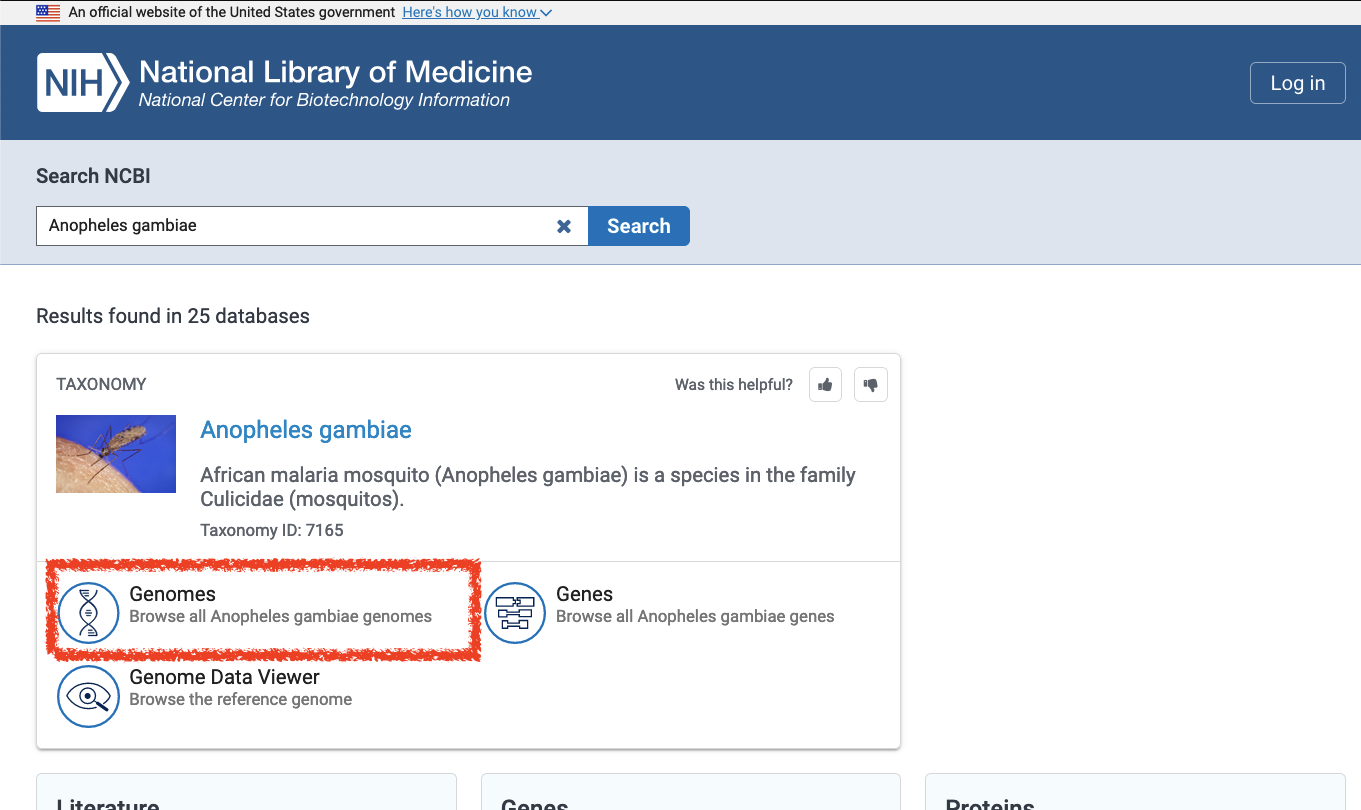
- Select the reference assembly at the top of the list; this entry is indicated with a green “reference genome” icon and a
GCF_identifer listed in the RefSeq column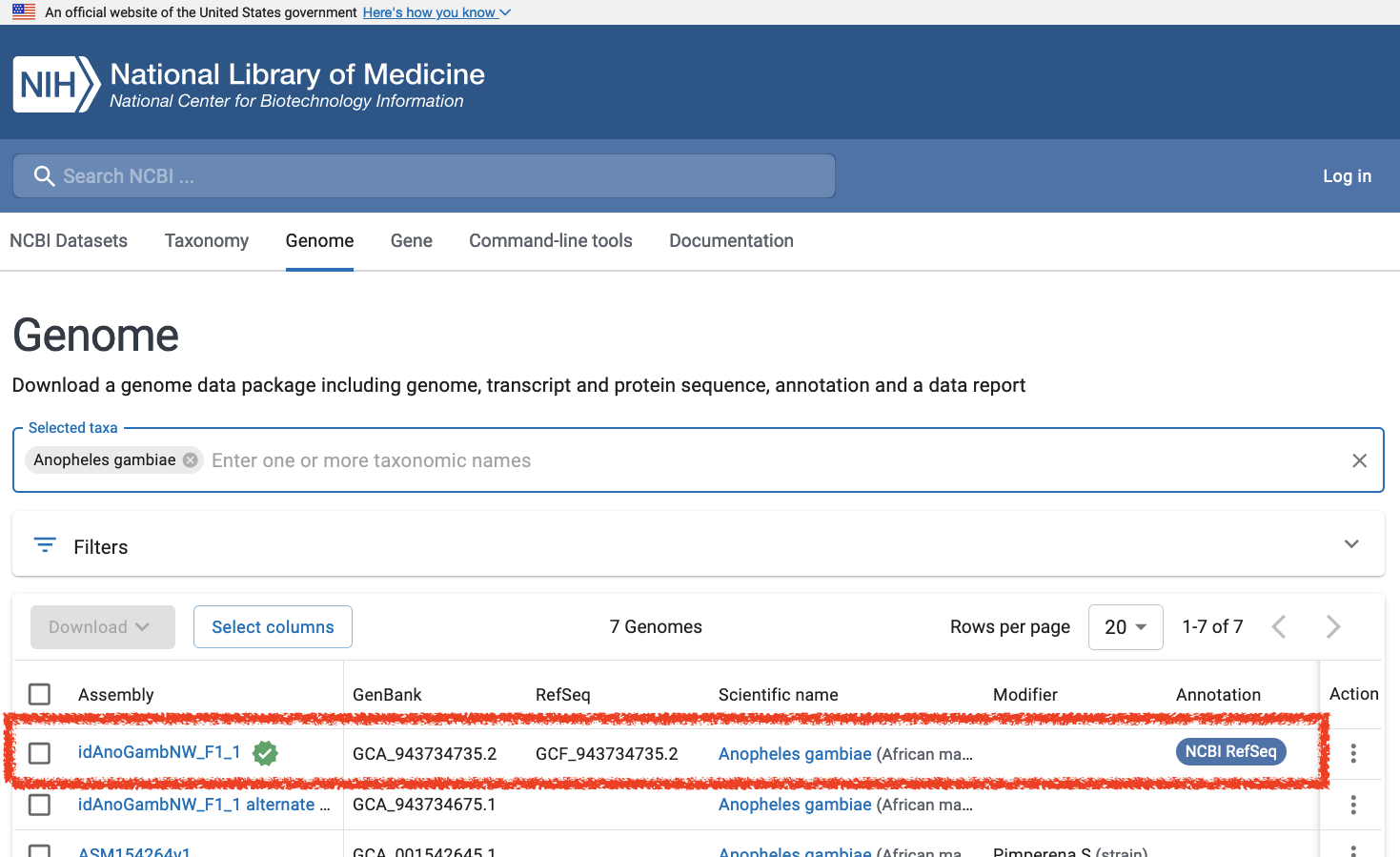
- Click on the FTP link
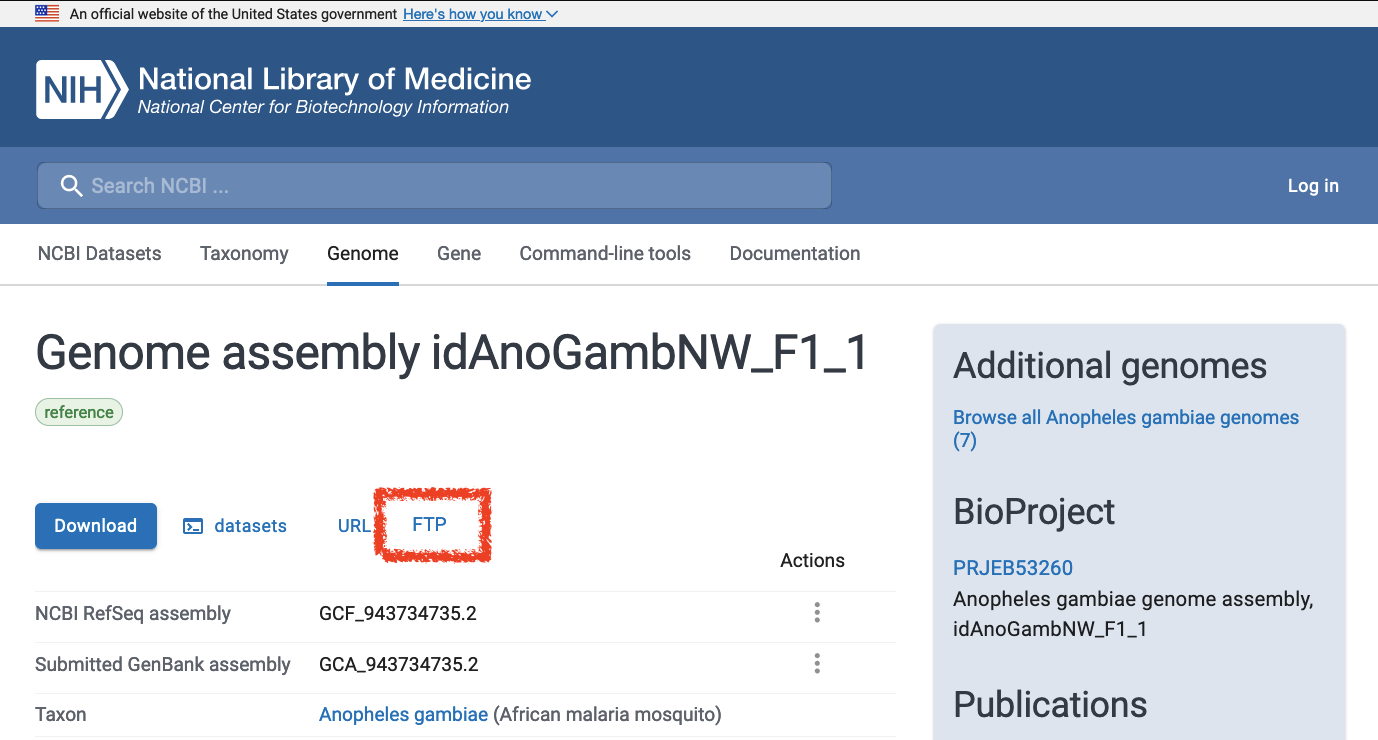
- Download the file with the suffix
gene_ontology.gaf.gz, e.g.GCF_943734735.2-RS_2023_12_gene_ontology.gaf.gz
3. If you cannot find annotations for your organism for download as described above
Get help from the GO helpdesk.
4. If your organism’s genome sequence is not yet publicly available
For example, if you have a set of new (protein) sequences that you want to annotate with GO terms, we recommend that you generate annotations using the latest version of InterProScan. For most genomic analyses, your input file should have one protein sequence per protein-coding gene, though any set of protein sequences can be used. Download InterProScan at https://www.ebi.ac.uk/interpro/about/interproscan.
More information on GO annotation formats
- GO has monthly releases
- Annotation files are taxon-specific, with a few exceptions including the Reactome and Candida Genome Database files
- Current format guides:
- GAF format 2.2
- GPAD + GPI companion files
Programmatic access to GO annotations
As for any resource from GO, GO annotations are accessible through the DOI-versioned release stored in Zenodo. + Please cite with a DOI and access the full bundle of the current release or any other archived release at Zenodo - record 1205166. DOI-versioned archives of each monthly GO release from 2018-08-09 are available through Zenodo; releases from 2004-03-01 to present are also available in our Archives.
Error or omission?
Any errors or omissions in annotations should be reported by writing to the GO helpdesk.